Can We
See Bonds with
X-Rays?
[Information in italics is not crucial to a
qualitative
understanding of what's going on. It is just added in case you start
thinking too deeply for the simple explanations.]
Table of Contents (clickable)
1) Why can't one use a
powerful light microscope to see bonds? 2) Just HOW is structural information
contained in diffracted X-rays?
a) A single pair of scattering points
b) An infinite row of evenly spaced
scattering points
c) A
row of evenly spaced pairs of scattering points
d) An hexagon of points ("benzene")
e) A pair of hexagons
f) A quartet of hexagons
g) A lattice of hexagons
h) Significance
3) What does Rosalind Franklin's x-ray
photo show about b-DNA?
a) Start by understanding the lightbulb filament.
b) Base-Pair Stacking
c) Diameter
d)
Double Helix - Major and Minor Grooves
4) What does the electron density in a
molecule look like? Are there Lewis Shared Pairs?
a) How to plot 3D electron density
b) What do e-density maps say about
molecules, atoms, and bonds?
c)
Accurate Difference Density maps show bonds
5) Conclusions
1) Why
can't one use a powerful light microscope to see
bonds?
Light is ELECTROMAGNETIC
RADIATION.
It consists of electric and magnetic fields that are oscillating
perpendicular to the direction in which the light is travelling (and
perpendicular to one another). For our present purposes we are
interested in the electric field, since that is the main actor in
pushing electrons around.
One can graph the oscillating electric field of light as a
sine wave, where the
vertical axis is
the FIELD STRENGTH (+
or - means
pointing one direction or the other, i.e. exerting
a force on
an electron in one directon or the other), and the horizontal axis
can be either TIME
or POSITION.
That is, we can plot
the change of the field with time at any given position (perhaps
where a particular atom is), or we can plot the change of field with
position along the direction of light propagation at some given
time.
|
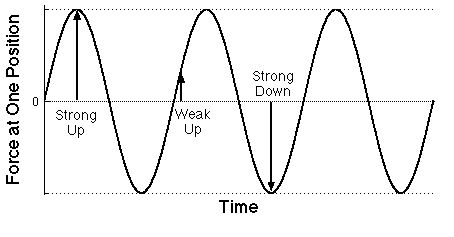
Graph of the force on an electron at some particular
position as a function of time due to the electric field of a
particular light wave. Of course the force on a proton would be just
the opposite. One over the time difference between
successive maxima is the frequency of the light.
| 
|
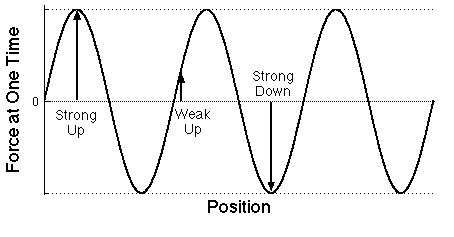
| Graph of the force on electrons at some
particular time as a function of the position of each
electron measured along the direction that the light wave is moving.
Note that only the labels have been changed from the plot above.
The distance between successive maxima is the wavelength
of the light. As for a water wave, there are two
different ways of describing light with a wave-shaped plot.
| 
|
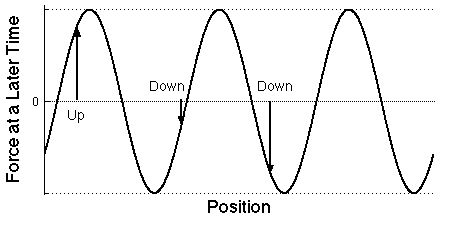
| Graph of the
force on the same electrons at a later time than
shown in the previous plot as a function of the position of each
electron position measured along the direction that the light wave is
moving. The forces can change in both size and direction. As time goes
by, the wave moves to the right, the direction of light propagation.
| 
|
The electric field of light pushes charged particles of
matter,
and the accelerating charges create new light waves that move off in
all directions, not just the direction in which the original light
wave is moving. This is called
SCATTERING. Of course
the electrons,
being more than 1000 times lighter than the lightest nucleus, are
accelerated much more than the nuclei, so
light
is scattered predominantly by electrons.
Thus if a light beam hits stuff and gets scattered, your eye
can
detect light even though it is not in position to be struck by the
incoming beam. Deflected light is the signal that there must have
been electrons in the light path to do the scattering.
It is good to have a way of knowing that
there are
electrons in the light path, but we want to know more. We want to
know where they are, how they are distributed, that
is we want
to know the STRUCTURE
of the matter that is
doing the
scattering. In particular, we want to know if electrons
are
gathered around nuclei to form atoms and whether pairs of electrons
concentrate between certain pairs of nuclei to constitute Lewis
shared-pair bonds. This is related to the problem of
RESOLUTION (how far
apart two things
have to be in order to tell that they are not a single piece).
One way to determine structure is to scan a VERY narrow beam
back
and forth, up and down across the sample and see where
scattering originates. This "raster "scanning (as in TV, or SPM) is
used in
scanning electron
microscopy and in an
SPM technique called SNOM (Scanning Near-field Optical Microscopy);
but beams can't be made narrow enough to resolve adjacent small
molecules, let alone atoms. Is there an alternative to scanning for
coding structural information into the scattered light? There must
be, because our eyes don't work by raster scanning.
|
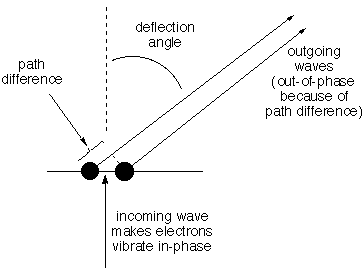
Structural information is coded into an INTERFERENCE PATTERN. Imagine
two electrons side-by-side in the path of the same incoming wave so
they are made to vibrate up and down in phase with one another.
Consider the oscillating electric field that hits a detector at a very
great distance [so distant that there is no significant difference
between the two electrons in the angle by which the beam is deflected
to hit a certain spot on the detector]. The total field is the sum of
the two scattered fields, and because the path for one is longer than
for the other, depending on the angle, the two waves will have
different phase at any given time,
one may be pushing up when the other is pushing down, for example. If
the path difference is exactly one (or two, or three...) waves, the
fields will reinforce one another to give a field twice as strong as
that from one electron. If the difference is half a wave, they will
cancel. Thus as the deflection angle increases from zero the amplitude
detected for the scattered wave will decrease, then increase, then
decrease, etc. How rapidly the
amplitude of the composite scattered beam changes with increasing angle
depends on the distance between the electrons and
the wavelength of the light. Decoding
this information about distance is the job of the lens of the
microscope (or the lens and retina of your eye). We need not be
concerned with the decoding, just with whether the structural
information is present to be decoded. |
|
[The "wave machine" shown in class illustrated how
such
interference arises.
To be
picky I should note that we're ignoring
certain pathological situations where
electrons in
different kinds of atoms may not vibrate precisely in phase with one
another even when they are being struck at the same time by the same
wave. This situation is called anomalous
dispersion, and can be quite useful.]
| If the wavelength is much greater
than the distance between the electrons, there can be no significant
difference in phase for any scattering angle, and
thus no clue in the scattered light that there are two separate
electrons. We can only resolve
objects whose separation is comparable to, or larger than, the light
wavelength. [Note that the greatest
phase difference occurs for light scattered at 90° from the incoming
ray. Thus to get maximum resolution from light of a given wavelength
one must collect scattered light up to a wide angle. This is why the
objective lens of a high-power microscope must get very close to the
object in view.] | 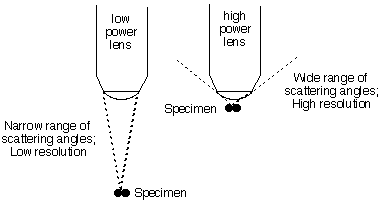
|
The shortest visible wavelength is
>400 nm = 4000 Å, ~ 2700
times the C-C bond distance. So forget using light microscopes to
resolve adjacent atoms within molecules or even adjacent molecules of
reasonable size.
If we want to resolve atoms, we must use waves that are about
1.5
Å (0.15 nm) long. This means
X-rays, but there is no
good lens for
focussing x-rays, so we must use a computer to decode the structural
information from the interference pattern collected over a wide range
of angles.
[High-kinetic-energy electrons also behave as waves
with short wavelength, and there are electric
"lenses" for them. They can be used for transmission electron
microscopy (TEM), but
they are much less generally useful than x-rays, because electrons are
so strongly scattered that they can't go through air or through more
than a few molecular layers of a sample.]
2) Just
HOW is structural information contained in diffracted
X-rays?
It is easy to appreciate that, if the wavelength of the light
is
short enough (x-rays), scattered light contains information about how
electrons are distributed in a sample. [A good analogy is a water
wave which encounters the pilings in a pier and generates a complex
pattern of reflected waves that depends on how the pilings were
arranged.]
The problem is how to decode the information.
Nowadays sophisticated computer programs can do the decoding
pretty easily for crystals of even rather large molecules - most
recently and dramatically (and at Yale) the ribosome with hundreds of
thousands of atoms. For most people these programs are just black
boxes. It is rewarding to think a little about how the information is
contained in the scattered X-rays. This is why we used the He/Ne
laser (wavelength 632.8 nm) in the lecture demonstration.
[First we think of scattering within a single plane,
as
visualized with the "wave machine". The first slides used in the
lecture demonstration consisted of vertical lines which, viewed from
above look like the "points" discussed below. I give this technical
information for the sake of honesty - you may stick with 2 dimensions
and think of points instead of lines to get the idea. It is cute that
in 3-D, just as a set of parallel lines on the slide give a row of
dots on the screen, a row of dots on the slide gives a set of
parallel lines on the screen.]
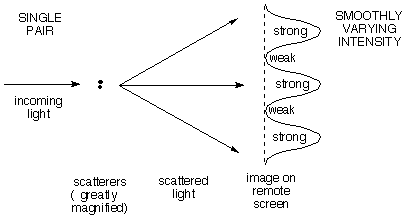
| a) A
single pair
of scattering points (e.g. electrons) gives an intensity
distribution that varies smoothly
with increasing scattering angle from maximum (straight
ahead) to minimum to maximum. The angular distance
between maxima (together with the wavelength) tells how far apart the
points are. Note the RECIPROCAL
relationship: the closer the points are together, the larger the
angular difference between scattered maxima. The first
deflected maximum comes when the wavelets from the two scatterers
differ by one wavelength, the second when they differ by two, and so on.
| 
|
| You are
probably familiar with this case already as the "two-slit" experiment
in which a plane wave of light travelling left to right passes through
two slits viewed end-on 1/4 and 3/4 up the left side of the diagram.
The resulting waves interfere (add to one another) to give at some
instant in time the pattern shown. The light areas are peaks and the
dark areas are troughs. Note that as the light moves far to the right
the patterns coalesce into a set of rays fanning out from a point
between the slits, and separated by gray areas of zero electric field.
This gives the smoothly varying intensity shown on the remote screen in
the diagram above. If you'd like to see an animation
of these interfering waves, click for the applet at the Physics 2000 website. |

|
| b) An infinite row
of evenly spaced scattering points gives a row of discrete
scattered spots. If the angle is chosen
so that the waves scattered from the first two points differ by exactly
one wavelength, the wave from the third will differ from that from the
first by two wavelengths, etc. So all the waves will be effectively in
phase to give very strong constructive interference. If the angle is
changed just a little bit so that the first two waves are not exactly
one wavelength different, the next will differ by twice as much, and so
on, so that ultimately cancellation will set in. |
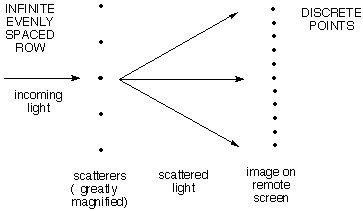
|
For example, if waves from the the first
two scatterers differed in
phase by only 1% of a wavelength, the 50th would differ from the
first by half a wavelength, and they would cancel. The 51st would
cancel the 2nd, and so on. So there is a lot of blank space between
successive maxima. As above, the angular distance between maxima
(together with the wavelength) tells how far apart adjacent points
are, and the relationship is reciprocal, large
spacing of
scatterers means close spacing of spots in the image.
The problem assigned in
class was to determine the spacing between the rows on the slide that
would give diffraction maxima separated by 10.8 cm on a screen at a
distance of 10.6 m when the wavelength is 633 nm. (the answer is about
65 μm). Doing the geometry will confirm your understanding of the
source of interference. Consider whether the observed points in the diffraction pattern should
be spaced exactly evenly.
|
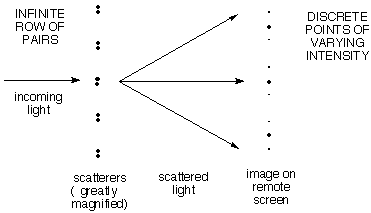
c)
More interesting is a row of evenly spaced PAIRS of
scattering points, where the pairs are closer together than
the repeat distance between pairs. Here
the pattern is the row of closely-spaced dots
expected from (b) for the large repeat distance
between pairs (remember the reciprocal relationship between spacing and
angle), but the intensities vary slowly as expected from (a) for the
smooth variation due to the members of a single closely-spaced pair.
|
|
Here's how to think about this:
The pattern which is to be repeated
generates a smooth variation of scattered intensity that tells about
the structure of the pattern. Here the pattern is a
pair of dots. We know above that any pair of dots with this spacing
will scatter with a smooth variation in intensity as shown in (a)
above. Since all of the pairs have the same spacing, they will all
scatter with the same angular dependence. Now all we have to worry
about is how the scattering from one pair interferes with that from the others.
The fact of the regular repetition allows one to observe this
scattered intensity only at the discrete positions allowed by the
repeat distance. [Importantly, the net scattered intensity is
unchanged. It is as if the intensity of the diffuse pattern is gathered
into the spots. This makes the spots easy to measure.]
This is like viewing the smooth distribution due to the
pattern through little holes whose spacing is determined by (and
reciprocal to) the row repeat, but the intensity is correspondingly amplified to preserve the total amount of scattered light.
|
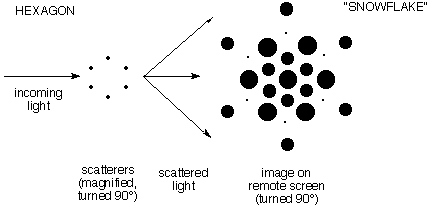
d)
The second demonstration showed scattering in three dimensions. It
involved a hexagon of points ("benzene"). A single
set of six points generated a smoothly varying "snowflake" pattern of
scattered intensity. The following illustration gives an idea of the
diffraction pattern, but fails to shows how smoothly it varies.
(Note: In this schematic illustration the
pictures of the scatterers and the diffraction image are turned 90° so you can see
them. Actually they lie on planes perpendicular to the ray of incoming
light.) Click here to see the Laser
Demonstration |
|
[In order to get enough
intensity to be visible the slide
actually had an enormous number of hexagons, all oriented the same
way but randomly placed. Randomly placed patterns give the same
diffraction as single patterns, but more intense. This contrasts with
a lattice of regularly placed patterns, which focuses the scattering
into discrete points, as described in e-g.]
|
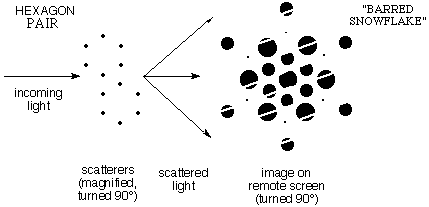
e)
When the pattern contained pairs of "benzenes" the diffraction showed
the same snowflake pattern but with intensity varying along the
direction in which the pairs are displaced. Click
here to see the Laser Demonstration
|  |
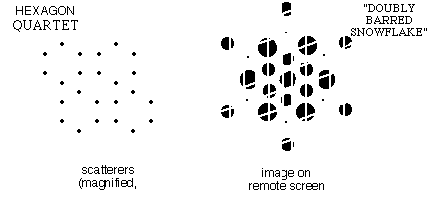
| f)
When the pattern contained quartets of "benzenes" at the corners of a
parallelogram, the same snowflake pattern was modulated in intensity in
two directions. Click here to see the Laser Demonstration |
|
g)
When all of the
"benzenes" in the slide fell on a regular lattice
with
standard row and column spacing, the diffraction pattern showed the
intensity distribution of the original snowflake but viewed only on
the two dimensional lattice of points allowed by the regular spacings
in rows and columns. This is the 2-dimensional analogue of
(c)
above. It is as if the snowflake were being viewed through
the
holes in a piece of pegboard.
Click
here to see the Laser Demonstration
h)
Significance:
The LOCAL ELECTRON
DISTRIBUTION
(molecular structure) is coded in the
INTENSITY of the spots
(in this case,
the underlying continuous snowflake pattern).
The SPACING OF THE ROWS
AND COLUMNS
(crystal lattice) is coded in the
ARRANGEMENT of the
spots (the "pegboard"
holes through which the snowflake is seen).
X-ray diffraction is usually measured with single crystals. The crystal
serves two purposes: First, it orients all of the
molecules in the same way, so the "snowflakes" reinforce. Second,
it concentrates all of the scattered light into a small number of
points which are bright enough to observe easily.
3) What
does Rosalind Franklin's x-ray photo show about b-DNA?
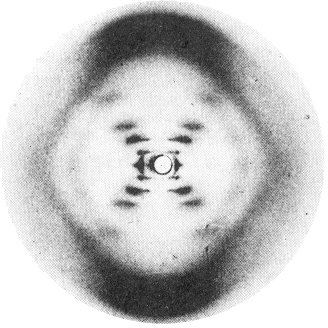
| The diffraction of He/Ne red light
by a slightly stretched filament from a broken lightbulb shows an "X"
with spots along each leg (see below). This dotted
"X" bears a strong resemblance to Franklin's photo (right) of DNA
fibers in which long molecules were oriented more or less vertically by
stretching. Since neither the filament nor the DNA
is crystalline, both show the rather diffuse pattern from an individual
object (like a pair - or hexagon - of dots) rather than the set of
sharp spots characteristic of an infinite regular lattice of such
objects. Of course there is a certain degree of
repetition within the helix itself, which gives rise to the features
that are discussed below. These helices are great examples of how
molecular structural information is present in the scattering pattern.
For example, the regular repetition from turn to turn along the helix
axis constitutes a repeating vertical pattern that gives rise to
horizontal bars, as in 2c or 2e above. |
|
a)
Start by understanding the lightbulb filament.
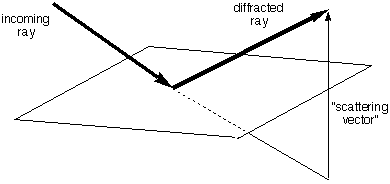
| We need the following rule:
All electrons on the
same
plane
perpendicular to the scattering vector
(the change in light direction, which bisects the incoming and outgoing rays, see figure) scatter
with the same phase.
[This is in fact how a mirror works - all
the silver atoms scatter light in phase when the incoming and outgoing
beam angles are the same.] For
purposes of calculating the scattered intensity, all the electron
density in such a plane could all be considered as concentrated at any
single point on the plane perpendicular to the scattering vector. THIS
IS A VERY NEAT TRICK. [It is easy
to prove this rule geometrically by showing that all the path lengths
(sum of incoming and outgoing) are the same]
| 
|
| So we can look for planes in the
sample that have a lot of electron density and think about the light
that is scattered (or "reflected") from them. For a
vertical helix (right) there is a lot of electron density on the
slanted planes shown (middle and far right), and not much on any given
parallel plane between them. Thus the indicated
scattering vectors should be imporant. That is, some of the light going
perpendicularly into the page should be deflected up to the left (and
down to the right) by the "planes" highlighted in the center figure.
Some should also be deflected down to the left (and up to the right) by
the "planes" highlighted in the right figure. These are the directions
of the arms of the "X". | 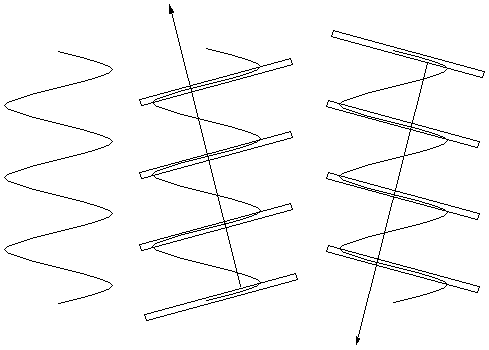 |
[Actually the helix must be slightly tilted
to bring these particular planes into reflection position. This may be
taken care of by rocking the sample a bit during exposure of the film,
or, as in the Franklin case, by the fact that different molecules in
the fiber have slightly different orientations so that many have the
desired orientation of slight tilt in any given direction. Even without
the axis tilting, there will be an analogous set of planes slightly
further along the helix (and a few degrees around to the right) that
will be tilted into the correct position. This is a fine point - don't
waste time on this if you don't see it right away. There are more
important
things to think about.]
For scattering in the indicated directions the e-density on
the
planes could be thought of as concentrated at the points where the
arrow intersects the planes. That is, there is scattering as if from
a row of dots, that we know would reinforce at a given succession of angles
(as in 2b above). There is very little electron density on the
interleaving planes, where it would have to be to cancel these
reinforcing waves.
Thus the "X" of dots in Franklin's photo
shows that DNA
is a helix, and the angle of the "X"
shows how tightly it
is wound (think about this).
Click
here to see the Laser Demonstration
b)
Base-Pair Stacking
Successive units of the DNA chain are spaced at a regular distance
along the axis of the helix. For example, one can see planes that
contain practically all of the carbon and nitrogen atoms of the
"bases" in stacked planes perpendicular to the axis. These planes are
much closer together than the "helix planes"
illustrated above
so they should reinforce for scattering at a much higher
angle. This gives rise to the very dark spots at the top and
bottom of the Franklin picture and shows that the stacking distance
is 3.4 Å (the thickness of an aromatic ring).
| c) Diameter
Much of the electron density of DNA is in the phosphorous and
oxygen atoms on the helical chain. Thus lots of the electron density
lies on the periphery of a cylinder. The illustration to the right
shows that planes that are nearly tangent to the cylinder contain lots
more electron density than those halfway between them (because the
center is not so dense in electrons. This gives lateral scattering that
measures the 20 Å diameter of the helix (the spots look like triangles
in Franklin's photo). | 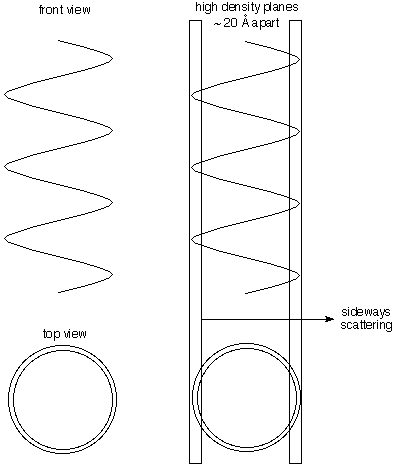 |
d)
Double Helix - Major and Minor Grooves
Other things being equal, spots get less intense as they move
to
higher scattering angle. This is observed as one moves out along the
arms of the "X" from the lightbulb filament scattering.
[This
is because the atoms have finite size, so that scattering from one
portion of an atom can interfere with that from another portion of the
same atom. This becomes more important as the relevant planes get
closer together for "high-angle" scattering.]
Things are different for DNA. As one moves out from the center
the
1st spot is rather weak, the 2nd and 3rd are strong, the 4th is too
weak to see, and the 5th is strong enough to see clearly - much
stronger than the fourth. What a queer sequence of
intensities!
[Incidentally, the undeflected beam would be
so strong as to wash out everything else - therefore a lead cup is put
in its path to intercept it, so there is no spot in the center of the
photograph.]
| The intensities show that DNA is a double
helix with a major and a minor groove
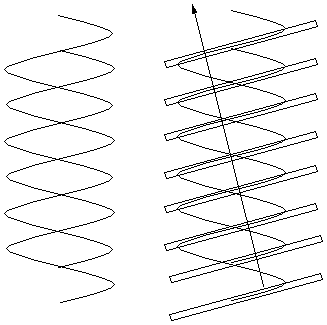
Had the structure involved an evenly spaced double
helix (as shown right) the spots on the "X" would be twice as
far apart (i.e. every other spot from the single helix would be
missing), because the slanted planes of electron density would be twice
as close together. Another way of saying
the same thing is the following: For a scattering angle where
waves from successive planes in the same helix differ by an odd number
of cycles (and thus reinforce one another), those from the other helix
differ from them by a half-integral number of cycles and cancel the
waves from the first helix. When reflections from
successive planes of a single helix differ by an even number of light
cycles, reflections from planes of the other helix differ from them by
an integral number of cycles and reinforce.
| 
|
| But
what if the helices are offset from even spacing to give major and
minor grooves as shown to the right? The pattern of
planes is like that of the spots in example
2c above - a widely spaced row of closely spaced pairs of
scatterers. The resulting diffraction pattern would also be the same -
a row of points spaced as for the row (single helix) spacing, but with
modulated intensities (weak, strong, strong, very weak, intermediate)
that can be used to measure the amount of offset between the two
helices. This is exactly what the Franklin photograph shows. |
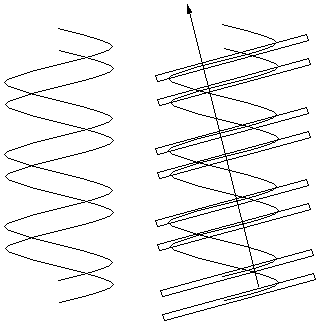
|
4) What
does the electron density in a molecule look like? Are there Lewis
Pairs?
a) How do you
visualize three dimensional plots
of electron density?
Electron density is a function of three variables - x, y, and
z
positions. How can you plot it?
On a 2-dimensional sheet of paper it is convenient to plot two
things against one another (two variables - independent and
dependent). It is harder to plot a function of two variables, e.g.
altitude (or temperature) as a function of latitude and longitude.
With color or a gray scale it is possible to plot such a function of
two variables. Another convention is to use a contour plot where
lines connect two-dimensional locations of a given value. You should
be familiar with such plots of isotherms (regions of the same
temperature) and with topographic maps showing a mountain as a series
of nested curves.(click
here for a brief exercise on 3D plots)
Plotting a function of three variables is much more difficult.
One
way would be to take a big block of clear gelatin and use an hypodermic
syringe with dye to inject more or less color in regions of certain
electron density. One could generate an image of such an object using
3-D computer graphics (this is precisely what Dean Dauger did to
represent the electron density in hydrogen-like atomic orbitals in
his program Atom in a Box, which we'll be using soon).
A practical
scheme for presentation on paper is to make a well-chosen cut through
3-D space to define a particular plane of interest (for example a
plane containing the atomic nuclei of a planar molecule) and then use
a contour plot to show electron density on that single plane. One
could make a lot a parallel planar cuts and show a bunch of such
contour graphs. No one said it would be easy. (here
is an example of such a planar map)
| The figure to the right shows
contours on part of an arbitrary slice through an e-density "map" from
an X-ray study of Rubofusarin. In the old days numbers giving the
e-density were typed on a page at the appropriate x,y positions by a
computer and the crystallographer then connected locations of the same
value with a pen to generate contours. This
was about 40 years ago - before it was easy and cheap to make computers
draw solid curves. There were no laser or ink jet printers or graphic
(or color) terminals. Electric typewriters or close relatives were the
common output devices. Imagine! From
examining the numbers it looks to me as though they were contouring at
values of 150, 200, 250, 300, 350, 400 (in arbitrary units, not e/Å3).
(from Stout and Jensen
"X-ray Structure Determination" 1968) |
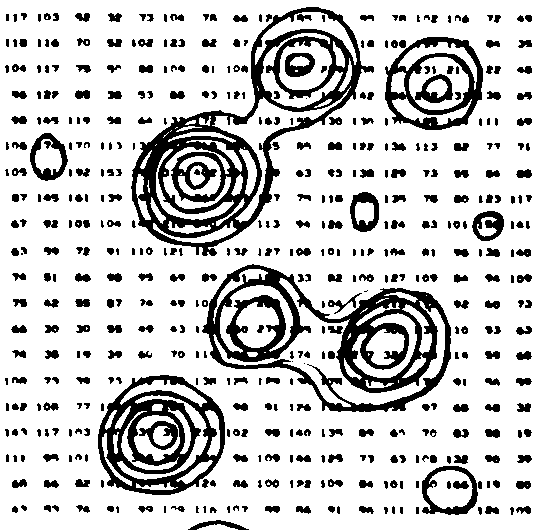
|
| Not all of the atoms fell on the
slice plotted above. Fortunately the molecule of
Rubofusarin is nearly planar, so the crystallographer could calculate
an analogous sheet of numbers for a different slice chosen to contain most of the
molecule's atoms. He then traced a similar set of
contours on an overlaid sheet of plain paper to give the figure shown
to the right. Here he drew contours at intervals of 1 e/Å3.
| 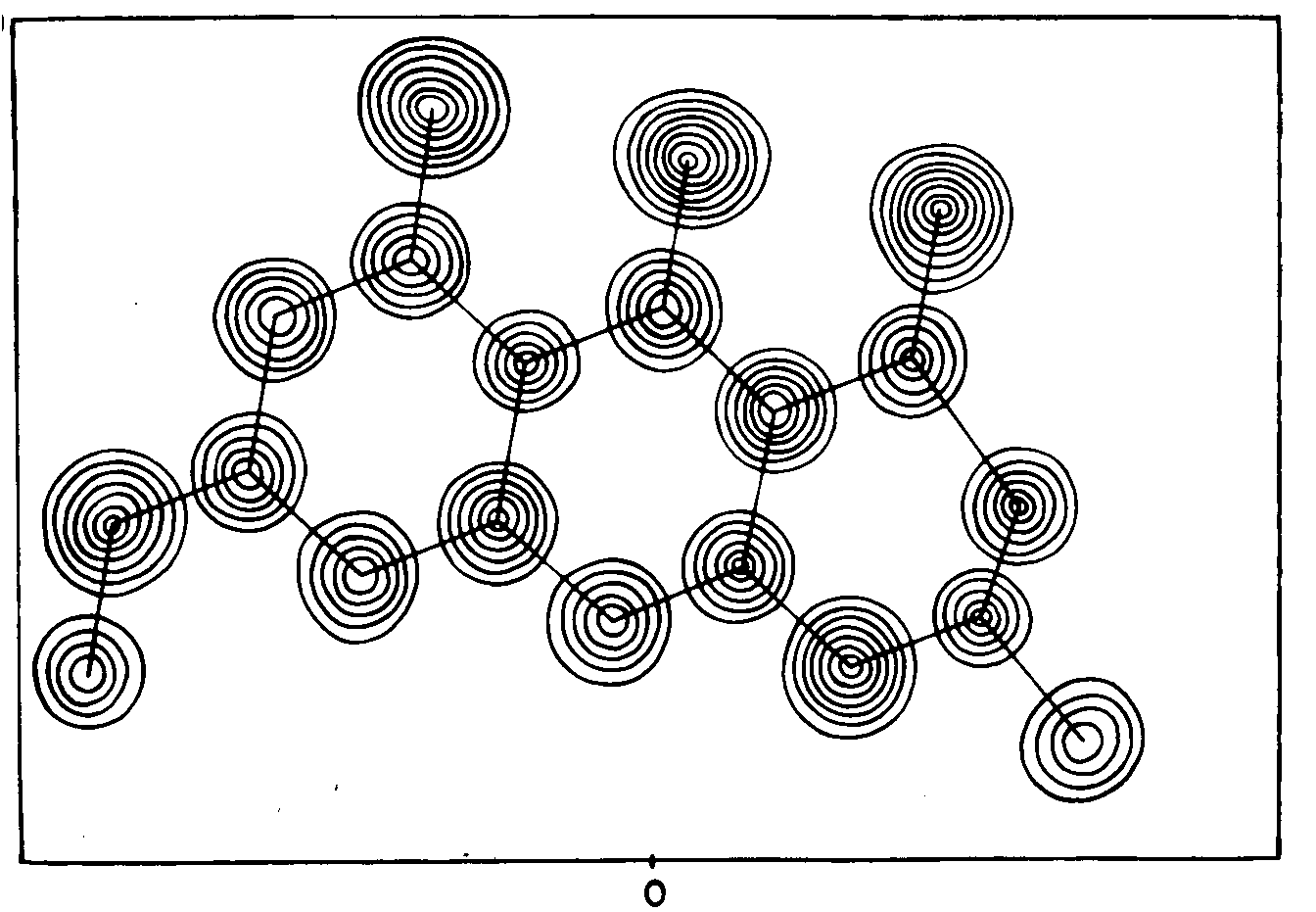 |
| Correspondence
of the spheres of high electron density with the non-hydrogen atoms in
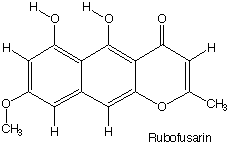
the formula of Rubofusarin is obvious.
| 
|
Rubofusarin has the advantage of being nearly planar, so that
a
single planar slice can give a good idea of the electron density of
almost all of its atoms. In a non-planar molecule one must choose
different slices to show features of the electron density with a
two-dimensional contour map. One approach, used in the 1940s in
connection with determination of the structure of the potassium salt
of penicillin by Dorothy
Crowfoot (later Hodgkin)
at Oxford, is to draw contour maps for a set of sequential, parallel
slices on transparent sheets and stack them up to give a
three-dimensional contour map, actually a sort of four-dimensional
graph! [KPenicillin
illustration]
b) What do
e-density maps say about molecules,
atoms, and bonds?
There is a lot of information in this e-density map.
Perhaps the most important is that this organic molecule looks like a collection of
spherical atoms with electron density increasing
toward the nucleus of each. There is certainly no
"double dot" of electron density between the atoms. In fact the spheres
don't even look distorted. There is no evidence from this
plot to support Lewis's idea that bonds consist of shared electron
pairs! Note that electron densities
identify the atoms:
hydrogens
have too little electron density to be visible in this map (less that 1
e/Å3)
only oxygen atoms achieve
electron density greater than 7 e/Å3 (seven
contour lines).
The nuclei
don't appear directly in the map, since they are too heavy to scatter
x-rays significantly, but one can guess that they are located where the
e-density is highest at the center of each sphere. [This
inference can be confirmed by experiments involving neutron scattering
- neutrons are scattered by nuclei and thus can reveal their position
directly.]
The bond lengths
show what kind of bond is involved:
| Carbon-Carbon |
Dist (Å) |
| Carbon-Oxygen |
Dist (Å) |
| Single
C-C
| 1.55
| |
Single C-O
| 1.37
|
| Double
C=C
| 1.32
| |
Double C=O
| 1.25
|
Such e-density maps confirm the reality of molecules and atoms
but
not of the Lewis shared-pair bonds.
c) Accurate
Difference Density maps show that
electrons DO shift (a little) to make bonds.
Of course one can wonder whether the apparently spherical
electron
density distributions that look so much like isolated atoms might be
at least a little bit distorted. The most sensitive way to check this
is with a "charge-density difference map". Instead of plotting the
total electron density, one plots the difference in density between
what is observed in very accurate experiments and what would be
expected for an analogous collection of perfectly spherical,
undistorted atoms. If there is no distortion upon forming bonds, the
difference density should be zero everywhere, but if electron density
shifts a little, there will be positive regions where density
accumulates and negative regions where it is depleted.
One lecture illustration showed such a difference density map for
a
molecule with single C-C bonds, aromatic C-C bonds, CN triple bonds,
and C-F single bonds. Solid contours show where electron density
increased upon forming bonds, and dashed (negative) contours show
where it was depleted. Notice that the amount of density shifted is
very modest - the contour level in the difference map is only 0.075
e/Å3, less than one tenth of the 1
e/Å3 interval used in contouring the total
e-density
map above. Even if the triple bond the maximum increase in e-density
between the nuclei is only 0.7 e/Å3, less that
10%
of the density near an oxygen nucleus. [Tetrafluorodinitrile illustration; see Lecture 6]
Lewis might have been gratified to see that, upon forming the
molecule, e-density accumulates between the atoms in "bonds", and in
an unshared pair on the nitrogen atom. [Lewis had died 20 years
before experiments of sufficient accuracy were possible.] It
is also satisfying to see that the greatest accumulation of e-density
is in the triple bond, followed by the "resonant" aromatic
"one-and-a-half" bonds, and the single bonds. [The much more
modest accumulation in the C-F bonds is surprising and requires some
mental gymnastics to rationalize. (see interview
with J. D. Dunitz)]
Perhaps even more striking is the cross sections
of the
bonds. The single and triple bonds have circular cross sections,
while the aromatic bonds have oval cross sections consistent with the
contribution from both sigma and pi bonds (which we will discuss a
lot more later). [Cumulene
overhead, see Lecture 6; see interview
with L.
Leiserowitz] [Cyclopropane illustration, see Lecture 7]
5)
Conclusions
So, using "x-ray vision", we can see
experimentally that
molecules
and atoms are real, and that bonding is associated with (modest)
accumulation of electrons between the bonded atoms.
Here is the answer to "How do you know?" - not because some
teacher or book told us there are atoms and bonds, but because it
seems plausible that previous scientists accurately reported x-ray
experiments of which we understand the significance. If we were
skeptical, we could repeat the experiments and see for ourselves.
Our next task is to understand WHY
the electrons gather into bonds and what properties bonds should have
(length, angles, strength, reactivity), and more fundamentally what
is special about octets, why there are electron shells, etc.
On
to Quantum Mechanics!
[Return
to Chem 125 Home Page]
latest revision 9/10/05
Comments on this page are welcomed by the author.
J. Michael McBride
Department
of
Chemistry, Yale
University
Box 208107, New Haven, CT 06520-8107
e-mail: j.mcbride@yale.edu
copyright
© 2001,2004,2005 J.M.McBride